The D-value (decimal reduction time) in microbiology indicates the time needed to reduce a microbial population by 90%. This signifies a 1 log reduction under specific conditions.
These conditions include a particular temperature or a particular disinfectant. It is a measure of a microorganism’s resistance to a sterilization or disinfection process.
Key Characteristics of D-value
- Logarithmic Reduction: A 90% reduction means reducing the microbial population to one-tenth of its initial value.
- Example: If you start with 1,000,000 cells (10610^6), a 90% reduction would leave 100,000 cells (10510^5).
- Specific to Conditions: The D-value depends on the:
- Type of microorganism.
- Environmental conditions (e.g., temperature, pH, or disinfectant used).
Importance of the D-value in microbiology
- It helps in determining the time required for effective sterilization or pasteurization processes.
- It ensures microbial safety in food processing by determining thermal or chemical treatment times.
- Validation: Used to validate disinfection and sterilization protocols.
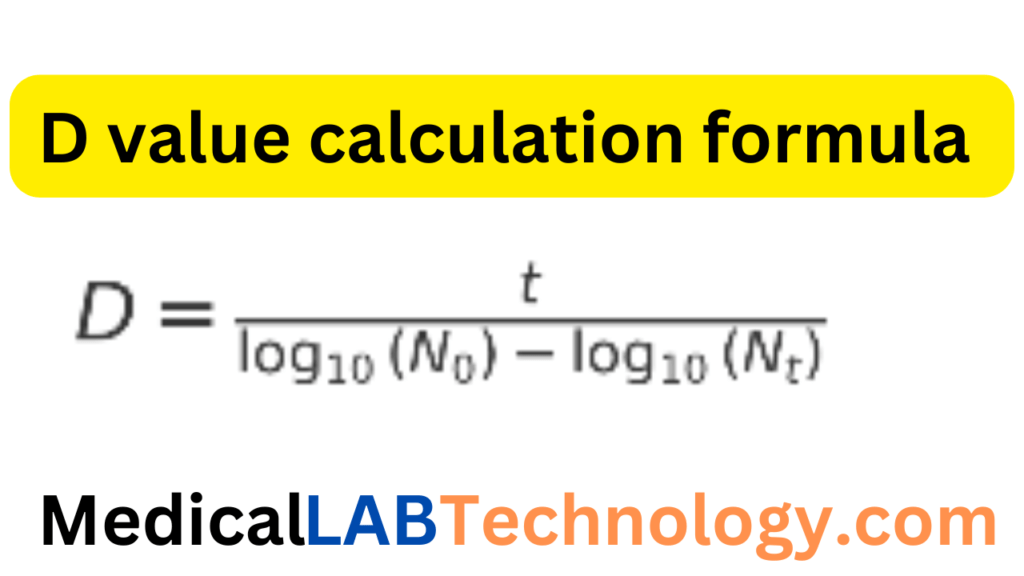
Formula of D value calculation
D=TimeLog ReductionD = \frac{\text{Time}}{\text{Log Reduction}}
Alternatively, from experimental data: D=1Slope of the Log Reduction CurveD = \frac{1}{\text{Slope of the Log Reduction Curve}}
Variants of D-value in microbiology
- D121D_{121}: D-value measured at 121°C, commonly used in autoclave sterilization.
- D-value with Disinfectants: Evaluates resistance of microbes to chemical agents.
How to calculate d value in microbiology
- First You need the microbial count (CFU/mL or another unit) at various times during the sterilization or disinfection process.
- Record the initial population (N0N_0) and the population (NtN_t) at specific time intervals.
- Convert the population data to a logarithmic scale:
- Log reduction=log10(N0)−log10(Nt)\text{Log reduction} = \log_{10}(N_0) – \log_{10}(N_t)
- Plot log10(Nt)\log_{10}(N_t) (y-axis) versus time (x-axis).
- The data should form a straight line if the reduction follows first-order kinetics.
- Find the slope (mm) of the line using the linear regression formula:
- m=Δ(log10(Nt))Δtimem = \frac{\Delta (\log_{10}(N_t))}{\Delta \text{time}}
- Calculate the D-Value: The D-value is the reciprocal of the absolute value of the slope: D=1∣m∣D = \frac{1}{|m|.
Explanation
- Suppose Initial population (N0N_0) = 10610^6
- Time intervals: 0, 1, 2, 3 minutes
- Populations (NtN_t): 106,105,104,10310^6, 10^5, 10^4, 10^3
Calculation
- log10(Nt)\log_{10}(N_t): 6,5,4,36, 5, 4, 3
- Slope (mm): m=6−30−3=−1m = \frac{6 – 3}{0 – 3} = -1
- D=1∣−1∣=1D = \frac{1}{|-1|} = 1 minute.
Key Notes:
- Specificity: D-values are temperature- or agent-specific.
- Units: The unit of the D-value matches the time intervals used (e.g., seconds, minutes).
- Verification: Ensure the linear relationship between log population and time holds true.
This method provides a standardized approach for assessing microbial resistance during sterilization or disinfection processes.